For some, it may be obvious / straight forward to implement, however the number of times I have been approached to ask for a script to transform the output of integer based fingerprints, such as MACCS keys or pharmacophore fingerprints coming out of MOE (the Molecular Operating Environment) into binary 000110101 representations so that molecules can be compared based on their Tanimoto similarity or some other measure or for input into a machine learning algorithm is astounding.
So here is a useful script that will convert integer based fingerprints into binary fingerprints.
import os, sys
class ConvertIntegerFPToBinary():
def __init__(self,descriptorFile,outputFile,label):
self.iFile = descriptorFile
self.oFile = outputFile
self.binaryFingerprints = []
self.integerList = []
self.label = label
def populateList(self,maxBitSize):
for i in range(1,maxBitSize+1):
self.integerList.append(str(i))
def convertData(self,maximumBitSize):
self.populateList(maximumBitSize)
inputFile = open(self.iFile,'r')
data = inputFile.readlines()
for i in range(1,len(data)):
splitdata = str(data[i]).replace("\"", "").split()
binaryFingerprint = []
for j in range(len(self.integerList)):
if self.integerList[j] in splitdata:
binaryFingerprint.append(1)
else:
binaryFingerprint.append(0)
binaryFingerprint.append(self.label)
self.binaryFingerprints.append(binaryFingerprint)
return self.binaryFingerprints
def postProcessing(self,fp):
binaryFingerprint = str(fp).replace("[", "").replace("]", "").replace("'", "")
return binaryFingerprint
def writeToFile(self):
outputFile = open(self.oFile,'w')
for i in range(len(self.binaryFingerprints)):
processedFingerprint = self.postProcessing(str(self.binaryFingerprints[i]))
if i == len(self.binaryFingerprints)-1:
outputFile.write(processedFingerprint)
else:
outputFile.write(processedFingerprint+"\n")
if __name__ == '__main__':
converter = ConvertIntegerFPToBinary(sys.argv[1],sys.argv[2],sys.argv[3])
binaryFingerprints = converter.convertData(166)
converter.writeToFile()
Saturday, 31 July 2010
Wednesday, 28 July 2010
Protection, Protection, Protection!

A long long time ago when I can still remember (well I think it was about 3 years ago), I set up a MediaWiki on my website (installation instructions are here). Whilst I was reaping the benefits of having a nice Web 2.0 interactive site, with ease of adding new content and web pages, something more sinister was lurking in the background. It was spam. Because there were few extensions at this time to prevent spam I thought it wouldn't happen to me. I was later told by my server administrator that I had a few gigs of spam on my wiki that was slowing down the whole server.
After removing the last wiki, I have now bravely decided to install it again (fortune favours the bold). This time round however, the first thing I have done is install all the possible extensions to combat spam. There is an exhaustive list of extensions here. There are a number of ways that one can restrict spam:
1. Restrict reading of the wiki, by setting $wgGroupPermissions['*']['read'] = false;
2. Prevent account creation, except by the administrator $wgGroupPermissions['*']['createaccount'] = false;
3. Restrict anonymous editing by setting $wgGroupPermissions['*']['edit'] = false;
4. Admin can also use the 'protect' facility to restrict access to certain pages.
5. Removing the login link / create account link using the following function:
function NoLoginLinkOnMainPage( &$personal_urls ){
unset( $personal_urls['login'] );
unset( $personal_urls['anonlogin'] );
return true;
}
$wgHooks['PersonalUrls'][]='NoLoginLinkOnMainPage';
6. Capatcha extension
The fact that edits have to be confirmed means that most bots will not be able to make edits to the wiki. This extension is simple to install, simply download the php file and add require_once( "$IP/extensions/ConfirmEdit/ConfirmEdit.php" ); to your localSettings.php file, like all other additions shown above.
Now that my wiki is much more secure I thought I would also add a few more extensions. Clearly it is useful to have your calendar displayed without going to say your gmail all the time. MediaWiki has an extension for this here. Just download the extension then go to this link to get details on your calendar and copy and paste all information between "googlecalendar" tags, ignoring all comments before the '?' and removing the "iframe" tag at the end.
I have also installed the chemistry extension, which works, but in my opinion is in rather a beta stage of development. It allows you to write chemical formulas into your wiki between "chemform" tags and it automatically corrects the sub and superscripting of atom numbers and charges. See here for examples. One of the problems however that I have found with this extension, is that say for example you have dioxygen with a single negative charge, and put
Thursday, 22 July 2010
Python makes the REST look easy
Web services are becoming an increasingly common phenomenon in the field of chemoinformatics, where data and services are now being published more openly on the internet. For those unfamiliar with the concept of web services, Wikipedia spells it out to be "application programming interfaces (API) or web APIs that are accessed via Hypertext Transfer Protocol (HTTP) and executed on a remote system hosting the requested services."
Traditionally people used SOAP web services, however they have waned in popularity relative to a new paradigm of web service called REST, which stands for Representational State Transfer. More information on RESTful web services can be found at the above link.
I would just like to illustrate with the use of some code, the simplicity of accessing some RESTful web services using Python. The web services we will be accessing are chemoinformatics web services running on a server at the University of Indiana under the CHEMBIOGrid projects.
The two services we will be accessing are:
1. 3D structure for a PubChem compound, retrieved from the Pub3D database
2. Generation of a 3D structure from a SMILES string, using the smi23d program
The code to access these services is shown below.
'''
Created on Jul 22, 2010
Program to access a REST web service hosted at Indiana University
@author: ed
'''
import urllib2, os, sys
class ChemoinformaticsRESTWebServices():
def __init__(self,url):
self.url = url
def callWS(self,parameter):
try:
url = self.url+parameter
data = urllib2.urlopen(url).read()
print data
except urllib2.HTTPError, e:
print "HTTP error: %d" % e.code
except urllib2.URLError, e:
print "Network error: %s" % e.reason.args[1]
if __name__ == '__main__':
#3D structure for a PubChem compound, retrieved from the Pub3D database
ws = ChemoinformaticsRESTWebServices("http://cheminfov.informatics.indiana.edu/rest/db/pub3d/")
ws.callWS(sys.argv[1])
ws.callWS(sys.argv[2])
#Generation of a 3D structure from a SMILES string, using the smi23d program
ws2 = ChemoinformaticsRESTWebServices("http://cheminfov.informatics.indiana.edu/rest/thread/d3.py/SMILES/")
ws2.callWS(sys.argv[3])
Traditionally people used SOAP web services, however they have waned in popularity relative to a new paradigm of web service called REST, which stands for Representational State Transfer. More information on RESTful web services can be found at the above link.
I would just like to illustrate with the use of some code, the simplicity of accessing some RESTful web services using Python. The web services we will be accessing are chemoinformatics web services running on a server at the University of Indiana under the CHEMBIOGrid projects.
The two services we will be accessing are:
1. 3D structure for a PubChem compound, retrieved from the Pub3D database
2. Generation of a 3D structure from a SMILES string, using the smi23d program
The code to access these services is shown below.
'''
Created on Jul 22, 2010
Program to access a REST web service hosted at Indiana University
@author: ed
'''
import urllib2, os, sys
class ChemoinformaticsRESTWebServices():
def __init__(self,url):
self.url = url
def callWS(self,parameter):
try:
url = self.url+parameter
data = urllib2.urlopen(url).read()
print data
except urllib2.HTTPError, e:
print "HTTP error: %d" % e.code
except urllib2.URLError, e:
print "Network error: %s" % e.reason.args[1]
if __name__ == '__main__':
#3D structure for a PubChem compound, retrieved from the Pub3D database
ws = ChemoinformaticsRESTWebServices("http://cheminfov.informatics.indiana.edu/rest/db/pub3d/")
ws.callWS(sys.argv[1])
ws.callWS(sys.argv[2])
#Generation of a 3D structure from a SMILES string, using the smi23d program
ws2 = ChemoinformaticsRESTWebServices("http://cheminfov.informatics.indiana.edu/rest/thread/d3.py/SMILES/")
ws2.callWS(sys.argv[3])
Wednesday, 21 July 2010
ベンゼン
Not sure of the title? Well if you knew Japanese you'd be ok. Since for some, or many of us, Japanese may not be the easiest of languages to pickup or understand, as an English speaker, I haven't got a clue what it means. Other than picking up a dictionary or attending some Japanese classes, you can use Google Translate. The advantage of Google Translate, is that it can convert between a multitude of different languages. If the suspense is killing you, the title actually means "Benzene".
I would be interested to see how well Google Translate works on other chemical names. If you're interested in converting say a batch load of names, there are Java and Python APIs available to access the web service.
Converting the name Benzene in English to its Romanian counterpart Benzen, works correctly.
Taking something more elaborate like: 13-amino-N-(2-{2-[(2-{[2-(2-aminoethyl)amino]ethyl}amino)ethyl]amino}ethyl)-
2,5,8,11-tetraazatridecanamide
returns -> 13-amino-N-(2 - (2 - [(2 - ([2 - (2-amino) etil] amino) amino) etil] amino) etil) -
2,5,8,11-tetraazatridecanamide
which I think is also right (with the exception of the loss in curly brace notation), though I will have to check.
Example Java code to translate
package eoc21;
import com.google.api.translate.Language;
import com.google.api.translate.Translate;
public class TranslateChemicalNames {
public static void main(String[] args) {
Translate.setHttpReferrer("en-fr");
try {
String translatedText = Translate.translate("tri-oxa-tri-silinane",
Language.ENGLISH, Language.ROMANIAN);
System.out.println(translatedText);
} catch (Exception ex) {
ex.printStackTrace();
}
}
}
I would be interested to see how well Google Translate works on other chemical names. If you're interested in converting say a batch load of names, there are Java and Python APIs available to access the web service.
Converting the name Benzene in English to its Romanian counterpart Benzen, works correctly.
Taking something more elaborate like: 13-amino-N-(2-{2-[(2-{[2-(2-aminoethyl)amino]ethyl}amino)ethyl]amino}ethyl)-
2,5,8,11-tetraazatridecanamide
returns -> 13-amino-N-(2 - (2 - [(2 - ([2 - (2-amino) etil] amino) amino) etil] amino) etil) -
2,5,8,11-tetraazatridecanamide
which I think is also right (with the exception of the loss in curly brace notation), though I will have to check.
Example Java code to translate
package eoc21;
import com.google.api.translate.Language;
import com.google.api.translate.Translate;
public class TranslateChemicalNames {
public static void main(String[] args) {
Translate.setHttpReferrer("en-fr");
try {
String translatedText = Translate.translate("tri-oxa-tri-silinane",
Language.ENGLISH, Language.ROMANIAN);
System.out.println(translatedText);
} catch (Exception ex) {
ex.printStackTrace();
}
}
}
Saturday, 17 July 2010
Ultrafast Shape Recognition
A few days ago, I was asked by a colleague who is working in the field of QSAR and virtual screening, to write an implementation of Pedro Ballester's Ultrafast Shape Recognition (USR) descriptor using the Python programming language.
Ballester's descriptor is a fast way to find molecules that closely resemble leads based on their shape. It has been shown to avoid the alignment problem, and to be up to 1500 times faster to calculate than other current methodologies. The shape descriptor makes the assumption that a molecule's shape can be uniquely defined by the relative position of its atoms and that three-dimensional shape can be characterised by one-dimensional distributions.
The source code along with HTML API documentation can be found on my github I hope this is of use to people. I have also uploaded a trial dataset of A42731 (Substance P Antagonists) and the resultant USR descriptor file.
Ballester's descriptor is a fast way to find molecules that closely resemble leads based on their shape. It has been shown to avoid the alignment problem, and to be up to 1500 times faster to calculate than other current methodologies. The shape descriptor makes the assumption that a molecule's shape can be uniquely defined by the relative position of its atoms and that three-dimensional shape can be characterised by one-dimensional distributions.
The source code along with HTML API documentation can be found on my github I hope this is of use to people. I have also uploaded a trial dataset of A42731 (Substance P Antagonists) and the resultant USR descriptor file.
Sunday, 11 July 2010
Designing a website using the Google Web Toolkit
Since I'm now going to be coding predominately in C++, I thought it would be nice to keep the Java going and give my website a new face. Whilst I'm not going to compare different JavaScript frameworks, I will give a few pointers as to why I chose the Google Web Toolkit (GWT).
- Firstly, I have previously developed a semantic web application using the GWT.
- Secondly, I have a stronger background in Java programming than Javascript. GWT allows you to write AJAX applications in Java and then compile the source to highly optimized JavaScript that runs across all browsers.
- Thirdly, because its Java I can use my favourtie IDE, Eclipse. Eclipse has a large number of tools to assist the developer, it also has the advantage that it comes with a GWT plugin, which can create projects seamlessly. Eclipse supports a number of version control systems (CVS, SVN, GitHub), necessary for any software project.
So if one were to develop a website or an online application, with GWT and Eclipse and support it under say the GitHub, how would one go about doing that?
Just follow these steps:
- Download Eclipse from http://www.eclipse.org/downloads/
- Add the GWT plugin. The appropriate urls are:
http://dl.google.com/eclipse/plugin/3.6
Eclipse 3.5 (Galileo)
http://dl.google.com/eclipse/plugin/3.5
Eclipse 3.4 (Ganymede)
http://dl.google.com/eclipse/plugin/3.4
Eclipse 3.3 (Europa)
http://dl.google.com/eclipse/plugin/3.3
3. Restart your Eclipse to make sure the workspace is refreshed.
4. To create a new GWT project, simply click on the GWT project icon in blue and white, or go to File-> New -> Project -> Google -> WebApplicationProject.
Click on the next button, then you will be prompted for a project name and package. You will also be given the option to specify the SDK you want to use in my case I have installed both the GWT -1.7.0 and GWT 2.0.4 versions. I would suggest upgrading to the newest version as from the GWT 2.0 mile stone, they no longer provide individual operating system .jar files. Previously I had a gwt-dev-linux.jar for my Ubuntu machine, however now the distributable .jar is called gwt-dev.jar.
This should produce a project structure similar to the one on my GitHub.
Code for the main class has been copied in here:
package eoc21.client;
import eoc21.shared.FieldVerifier;
import com.google.gwt.core.client.EntryPoint;
import com.google.gwt.core.client.GWT;
import com.google.gwt.event.dom.client.ClickEvent;
import com.google.gwt.event.dom.client.ClickHandler;
import com.google.gwt.event.dom.client.KeyCodes;
import com.google.gwt.event.dom.client.KeyUpEvent;
import com.google.gwt.event.dom.client.KeyUpHandler;
import com.google.gwt.user.client.rpc.AsyncCallback;
import com.google.gwt.user.client.ui.Button;
import com.google.gwt.user.client.ui.DialogBox;
import com.google.gwt.user.client.ui.HTML;
import com.google.gwt.user.client.ui.Label;
import com.google.gwt.user.client.ui.RootPanel;
import com.google.gwt.user.client.ui.TextBox;
import com.google.gwt.user.client.ui.VerticalPanel;
/**
* Entry point classes define
onModuleLoad().*/
public class GWTWebSite implements EntryPoint {
/**
* The message displayed to the user when the server cannot be reached or
* returns an error.
*/
private static final String SERVER_ERROR = "An error occurred while "
+ "attempting to contact the server. Please check your network "
+ "connection and try again.";
/**
* Create a remote service proxy to talk to the server-side Greeting service.
*/
private final GreetingServiceAsync greetingService = GWT
.create(GreetingService.class);
/**
* This is the entry point method.
*/
public void onModuleLoad() {
final Button sendButton = new Button("Send");
final TextBox nameField = new TextBox();
nameField.setText("GWT User");
final Label errorLabel = new Label();
// We can add style names to widgets
sendButton.addStyleName("sendButton");
// Add the nameField and sendButton to the RootPanel
// Use RootPanel.get() to get the entire body element
RootPanel.get("nameFieldContainer").add(nameField);
RootPanel.get("sendButtonContainer").add(sendButton);
RootPanel.get("errorLabelContainer").add(errorLabel);
// Focus the cursor on the name field when the app loads
nameField.setFocus(true);
nameField.selectAll();
// Create the popup dialog box
final DialogBox dialogBox = new DialogBox();
dialogBox.setText("Remote Procedure Call");
dialogBox.setAnimationEnabled(true);
final Button closeButton = new Button("Close");
// We can set the id of a widget by accessing its Element
closeButton.getElement().setId("closeButton");
final Label textToServerLabel = new Label();
final HTML serverResponseLabel = new HTML();
VerticalPanel dialogVPanel = new VerticalPanel();
dialogVPanel.addStyleName("dialogVPanel");
dialogVPanel.add(new HTML("Sending name to the server:"));
dialogVPanel.add(textToServerLabel);
dialogVPanel.add(new HTML("
Server replies:"));
dialogVPanel.add(serverResponseLabel);
dialogVPanel.setHorizontalAlignment(VerticalPanel.ALIGN_RIGHT);
dialogVPanel.add(closeButton);
dialogBox.setWidget(dialogVPanel);
// Add a handler to close the DialogBox
closeButton.addClickHandler(new ClickHandler() {
public void onClick(ClickEvent event) {
dialogBox.hide();
sendButton.setEnabled(true);
sendButton.setFocus(true);
}
});
// Create a handler for the sendButton and nameField
class MyHandler implements ClickHandler, KeyUpHandler {
/**
* Fired when the user clicks on the sendButton.
*/
public void onClick(ClickEvent event) {
sendNameToServer();
}
/**
* Fired when the user types in the nameField.
*/
public void onKeyUp(KeyUpEvent event) {
if (event.getNativeKeyCode() == KeyCodes.KEY_ENTER) {
sendNameToServer();
}
}
/**
* Send the name from the nameField to the server and wait for a response.
*/
private void sendNameToServer() {
// First, we validate the input.
errorLabel.setText("");
String textToServer = nameField.getText();
if (!FieldVerifier.isValidName(textToServer)) {
errorLabel.setText("Please enter at least four characters");
return;
}
// Then, we send the input to the server.
sendButton.setEnabled(false);
textToServerLabel.setText(textToServer);
serverResponseLabel.setText("");
greetingService.greetServer(textToServer,
new AsyncCallback
public void onFailure(Throwable caught) {
// Show the RPC error message to the user
dialogBox
.setText("Remote Procedure Call - Failure");
serverResponseLabel
.addStyleName("serverResponseLabelError");
serverResponseLabel.setHTML(SERVER_ERROR);
dialogBox.center();
closeButton.setFocus(true);
}
public void onSuccess(String result) {
dialogBox.setText("Remote Procedure Call");
serverResponseLabel
.removeStyleName("serverResponseLabelError");
serverResponseLabel.setHTML(result);
dialogBox.center();
closeButton.setFocus(true);
}
});
}
}
// Add a handler to send the name to the server
MyHandler handler = new MyHandler();
sendButton.addClickHandler(handler);
nameField.addKeyUpHandler(handler);
}
}
Essentially we create a class for the entry point for the website (i.e. home), we implement the onModuleLoad() method. The tutorial defines a number of widgets: a button called "send", a TextBox with a default value "GWT", the text box and button are added to the root panel. A DialogBox is then created that will display the results. A close button is added to the DialogBox with a click handler to close the DialogBox if a user clicks on the close button.
A nested class called MyHandler is constructed that implements both the ClickHandler and KeyUpHandler interfaces, which has to implement the onClick() and onKeyUpUp() methods. If a user clicks the send button or types in information in the textbox, it is sent to the server, via the sendNameToServer() method, which implements an asynchronous remote proceedual call to the server.
5. Get Egit
You're probably now wondering how I integrated Eclipse with GitHub? Well, there is also a plugin to allow you to push your code to GitHub. The plugin is called Egit, and can be downloaded from: http://download.eclipse.org/egit/updates. A full and more comprehensive tutorial on Egit can be found at http://www.vogella.de/articles/EGit/article.html.
6. Mavenize and manage those dependencies
If you're new to Java, or you haven't heard of Maven, you should read this (http://maven.apache.org/). Apache Maven is a software project management and comprehension tool. Based on the concept of a project object model (POM), Maven can manage a project's build, reporting and documentation from a central piece of information. I will talk about maven, and mavenizing a GWT project in another post.
GWT Simple Demo
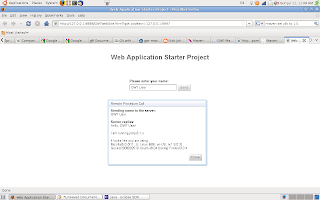
The result of our simple GWT application / website demo is shown in the image below. On clicking the send button an RPC (remote proceedual call) is made, the server responds with "Sending name to the server"+ your input text, then it replies with "hello" and the name of the user and details of the operating system and browser used (in this case Ubuntu 8.0.4 and FireFox).
Tuesday, 6 July 2010
Name2Structure and Back Again, A New Adventure
A recent presentation I found on line, titled "Chemical Information Mining: Possibilities and Pitfalls", attracted my attention. It was given by Neil Stutchbury on the 20th of May 2009 at the CICAG (Chemical Information and Computer Applications Group) Scientific Text and Data Mining conference in Burlington House London. The presentation covered a number of key points, firstly extracting chemical entities in text, secondly converting chemical names to structures, thirdly a user survey and finally points for future possible extensions.
Since I have now accepted a position at OpenEye, to primarily work on Lexichem, I am particularly interested in his discussion on name to structure. You're probably wondering what comes under the umbrella of name? Well there are a number of ever emerging types of names, too many to list in this blog post. However the ones currently under discussion are: trade names, common names, natural products, abbreviations, molecular formulae and IUPAC names. A trade name according to wikipedia, is "the name which a business trades under for commercial purposes", an example of usage would be "Aspirin"
which is dissimilar to its chemical name "acetylsalicylic acid". A common name, is a name in general use in a community, caffiene is a common name for 1,3,7-trimethyl-1H-purine- 2,6(3H,7H)-dione. Natural products are considered chemical compounds or substances produced by a living organism such as Taxol. In chemistry, a number of abbreviations are used to shorten the form of a word or phrase, examples include DMSO - dimethyl sulfoxide and THF - tetrahydrofuran. People in everyday life often talk about ordering H2O at the bar after a heavy night out, H20 is the formula for water.
IUPAC names are based on IUPAC nomenclature, a system of naming chemical compounds. It is developed and kept up to date by the International Union of Pure and Applied Chemistry (IUPAC). There are currently two books detailing rules for naming organic and inorganic compounds, which will be the subject of a future blog post.
When starting a new position, in any field, it is always interesting to know the competition. So who (both open and commerical) produces name to structure tools? Well, the (main) suppliers are shown in the table below:
University of Cambridge ->OSCAR
ChemAxon -> Chemicalize.org
InfoChem -> Annotator and ICN2S
ChemMantis -> SureChem (NER) with ACD/Name; ACD/NTS / Batch (N2S)
CambridgeSoft ->Name=Struct
OpenEye ->Lexichem TK
Accelrys ->ChemMining
TEMIS/MDL ->Chemical Entity Relationships Skill Cartridge
MPirics ->Chemical Content Recognition
OSCAR
Having first hand experience, coming from the University of Cambridge and Peter Murray-Rust's group, I can recommend OPSIN (the N2S module), which is open source code, and written in Java. Whilst OPSIN is showing progress, it is still in its infancy.
Chemicalize.org
A product of ChemAxon, is hosted online. They have used the jQuery javascript library to create a form that a user can paste in chemical names or upload a small (4KB) file to convert names to structure. It should also be noted however that ChemAxon have their own Java API to convert names to structure and structures to names.
A simple program to read in molecules, then convert the molecules to IUPAC and traditional names is shown.
package eoc21;
import java.io.IOException;
import chemaxon.formats.MolFormatException;
import chemaxon.formats.MolImporter;
import chemaxon.license.LicenseManager;
import chemaxon.license.LicenseProcessingException;
import chemaxon.marvin.calculations.IUPACNamingPlugin;
import chemaxon.marvin.plugin.PluginException;
import chemaxon.struc.Molecule;
/**
* Converts structure to name using ChemAxon's java API.
* @author ed
*
*/
public class Name2Structure {
public static void main(String[] args) throws PluginException, MolFormatException, IOException, LicenseProcessingException {
LicenseManager.setLicenseFile("license.cxl");
MolImporter mi = new MolImporter("150mols.smi");
Molecule mol;
IUPACNamingPlugin plugin = new IUPACNamingPlugin();
while((mol = mi.read()) != null){
plugin.setMolecule(mol);
plugin.run();
String preferredIUPACName = plugin.getPreferredIUPACName();
String traditionalName = plugin.getTraditionalName();
System.out.println(preferredIUPACName);
}
mi.close();
}
}
Annotator
This product can extract chemically relevant entities from text and convert the names to structure. It supports: systematic names, trivial names, trade names as well as inchis and CAS Registry Numbers.
ChemMantis
An integrated system for chemistry-based entity extraction and document mark-up enabling access to the rich resource of online chemistry know as ChemSpider.
Name=Struct
CambridgeSoft supports both name to structure and structure to name conversions. It offers support for charged compounds and salts, highly symmetric structures and many other types of inorganic and organometallics.
Lexichem TK
A C++ toolkit provided by OpenEye to convert names to structure, structure to names and has the ability to convert the names into different languages such as Japanese, Romanian and Hungarian. Example of name to structure code taken from the Lexichem documentation is given below:
***********************************************************************/
#include "openeye.h"
#include "oeplatform.h"
#include "oesystem.h"
#include "oechem.h"
#include "oeiupac.h"
#include "nam2mol_example.itf"
using namespace OEPlatform;
using namespace OESystem;
using namespace OEChem;
using namespace OEIUPAC;
using namespace std;
#ifndef STDIN_FILENO
#define STDIN_FILENO 0
#endif
int main(int argc, char *argv[])
{
OESetMemPoolMode(OEMemPoolMode::SingleThreaded|OEMemPoolMode::UnboundedCache);
OEThrow.Info("Lexichem nam2mol example");
OEThrow.Info(" Lexichem version: %s", OEIUPACGetRelease());
OEInterface itf(InterfaceData, argc, argv);
oeifstream infile;
string inname=itf.Get("-in");
if (inname=="-")
{
if (!infile.openfd(STDIN_FILENO, true)) // read from stdin
OEThrow.Fatal("Unable to read from stdin");
}
else
{
if (!infile.open(inname))
OEThrow.Fatal("Unable to open input file: %s\n", inname.c_str());
}
oemolostream outfile;
if (!outfile.open(itf.Get("-out")))
OEThrow.Fatal("Unable to create output file: %s\n",
itf.Get("-out").c_str());
unsigned int language = OEGetIUPACLanguage(itf.Get("-language"));
OEGraphMol mol;
char buffer[8192];
bool done;
while (infile.getline(buffer,8192))
{
mol.Clear();
// Speculatively reorder CAS permuted index names
std::string str = OEReorderIndexName(buffer);
if (str.empty()) str = buffer;
if (language != OELanguage::AMERICAN)
{
str = OEFromUTF8(str.c_str());
str = OELowerCaseName(str.c_str());
str = OEFromLanguage(str.c_str(),language);
}
done = OEParseIUPACName(mol,str.c_str());
if (!done && itf.Get("-empty"))
{
mol.Clear();
done = true;
}
if (done)
{
if (itf.Has("-tag"))
OESetSDData(mol,itf.Get("-tag"),buffer);
mol.SetTitle(buffer);
OEWriteMolecule(outfile,mol);
}
}
return 0;
}
ChemMining
This software has the advantage, that it can be incorporated into Pipeline Pilot workflows. ChemMining can extract chemical entities from documents and includes: chemical names, IUPAC names, SMILES strings and common and brand names.
Chemical Entity Relationships Skill Cartridge
Developed by January 2006, this cartridge identifies chemical compound names, chemical classes and molecular formulae in text documents and translates extracted information into the chemist’s language: the chemical structure. The software includes the ability to identify chemical terms and assign them to specific chemical concepts according to semantic categories (another area of interest, the semantic web and chemistry, which I will blog about later in the year).
Chemical Content Recognition
This is a service delivered by MPirics. It allows chemists to chemical entities from text, the main goal being patents.
Since I have now accepted a position at OpenEye, to primarily work on Lexichem, I am particularly interested in his discussion on name to structure. You're probably wondering what comes under the umbrella of name? Well there are a number of ever emerging types of names, too many to list in this blog post. However the ones currently under discussion are: trade names, common names, natural products, abbreviations, molecular formulae and IUPAC names. A trade name according to wikipedia, is "the name which a business trades under for commercial purposes", an example of usage would be "Aspirin"
which is dissimilar to its chemical name "acetylsalicylic acid". A common name, is a name in general use in a community, caffiene is a common name for 1,3,7-trimethyl-1H-purine- 2,6(3H,7H)-dione. Natural products are considered chemical compounds or substances produced by a living organism such as Taxol. In chemistry, a number of abbreviations are used to shorten the form of a word or phrase, examples include DMSO - dimethyl sulfoxide and THF - tetrahydrofuran. People in everyday life often talk about ordering H2O at the bar after a heavy night out, H20 is the formula for water.
IUPAC names are based on IUPAC nomenclature, a system of naming chemical compounds. It is developed and kept up to date by the International Union of Pure and Applied Chemistry (IUPAC). There are currently two books detailing rules for naming organic and inorganic compounds, which will be the subject of a future blog post.
When starting a new position, in any field, it is always interesting to know the competition. So who (both open and commerical) produces name to structure tools? Well, the (main) suppliers are shown in the table below:
University of Cambridge ->OSCAR
ChemAxon -> Chemicalize.org
InfoChem -> Annotator and ICN2S
ChemMantis -> SureChem (NER) with ACD/Name; ACD/NTS / Batch (N2S)
CambridgeSoft ->Name=Struct
OpenEye ->Lexichem TK
Accelrys ->ChemMining
TEMIS/MDL ->Chemical Entity Relationships Skill Cartridge
MPirics ->Chemical Content Recognition
OSCAR
Having first hand experience, coming from the University of Cambridge and Peter Murray-Rust's group, I can recommend OPSIN (the N2S module), which is open source code, and written in Java. Whilst OPSIN is showing progress, it is still in its infancy.
Chemicalize.org
A product of ChemAxon, is hosted online. They have used the jQuery javascript library to create a form that a user can paste in chemical names or upload a small (4KB) file to convert names to structure. It should also be noted however that ChemAxon have their own Java API to convert names to structure and structures to names.
A simple program to read in molecules, then convert the molecules to IUPAC and traditional names is shown.
package eoc21;
import java.io.IOException;
import chemaxon.formats.MolFormatException;
import chemaxon.formats.MolImporter;
import chemaxon.license.LicenseManager;
import chemaxon.license.LicenseProcessingException;
import chemaxon.marvin.calculations.IUPACNamingPlugin;
import chemaxon.marvin.plugin.PluginException;
import chemaxon.struc.Molecule;
/**
* Converts structure to name using ChemAxon's java API.
* @author ed
*
*/
public class Name2Structure {
public static void main(String[] args) throws PluginException, MolFormatException, IOException, LicenseProcessingException {
LicenseManager.setLicenseFile("license.cxl");
MolImporter mi = new MolImporter("150mols.smi");
Molecule mol;
IUPACNamingPlugin plugin = new IUPACNamingPlugin();
while((mol = mi.read()) != null){
plugin.setMolecule(mol);
plugin.run();
String preferredIUPACName = plugin.getPreferredIUPACName();
String traditionalName = plugin.getTraditionalName();
System.out.println(preferredIUPACName);
}
mi.close();
}
}
Annotator
This product can extract chemically relevant entities from text and convert the names to structure. It supports: systematic names, trivial names, trade names as well as inchis and CAS Registry Numbers.
ChemMantis
An integrated system for chemistry-based entity extraction and document mark-up enabling access to the rich resource of online chemistry know as ChemSpider.
Name=Struct
CambridgeSoft supports both name to structure and structure to name conversions. It offers support for charged compounds and salts, highly symmetric structures and many other types of inorganic and organometallics.
Lexichem TK
A C++ toolkit provided by OpenEye to convert names to structure, structure to names and has the ability to convert the names into different languages such as Japanese, Romanian and Hungarian. Example of name to structure code taken from the Lexichem documentation is given below:
***********************************************************************/
#include "openeye.h"
#include "oeplatform.h"
#include "oesystem.h"
#include "oechem.h"
#include "oeiupac.h"
#include "nam2mol_example.itf"
using namespace OEPlatform;
using namespace OESystem;
using namespace OEChem;
using namespace OEIUPAC;
using namespace std;
#ifndef STDIN_FILENO
#define STDIN_FILENO 0
#endif
int main(int argc, char *argv[])
{
OESetMemPoolMode(OEMemPoolMode::SingleThreaded|OEMemPoolMode::UnboundedCache);
OEThrow.Info("Lexichem nam2mol example");
OEThrow.Info(" Lexichem version: %s", OEIUPACGetRelease());
OEInterface itf(InterfaceData, argc, argv);
oeifstream infile;
string inname=itf.Get
if (inname=="-")
{
if (!infile.openfd(STDIN_FILENO, true)) // read from stdin
OEThrow.Fatal("Unable to read from stdin");
}
else
{
if (!infile.open(inname))
OEThrow.Fatal("Unable to open input file: %s\n", inname.c_str());
}
oemolostream outfile;
if (!outfile.open(itf.Get
OEThrow.Fatal("Unable to create output file: %s\n",
itf.Get
unsigned int language = OEGetIUPACLanguage(itf.Get
OEGraphMol mol;
char buffer[8192];
bool done;
while (infile.getline(buffer,8192))
{
mol.Clear();
// Speculatively reorder CAS permuted index names
std::string str = OEReorderIndexName(buffer);
if (str.empty()) str = buffer;
if (language != OELanguage::AMERICAN)
{
str = OEFromUTF8(str.c_str());
str = OELowerCaseName(str.c_str());
str = OEFromLanguage(str.c_str(),language);
}
done = OEParseIUPACName(mol,str.c_str());
if (!done && itf.Get
{
mol.Clear();
done = true;
}
if (done)
{
if (itf.Has
OESetSDData(mol,itf.Get
mol.SetTitle(buffer);
OEWriteMolecule(outfile,mol);
}
}
return 0;
}
ChemMining
This software has the advantage, that it can be incorporated into Pipeline Pilot workflows. ChemMining can extract chemical entities from documents and includes: chemical names, IUPAC names, SMILES strings and common and brand names.
Chemical Entity Relationships Skill Cartridge
Developed by January 2006, this cartridge identifies chemical compound names, chemical classes and molecular formulae in text documents and translates extracted information into the chemist’s language: the chemical structure. The software includes the ability to identify chemical terms and assign them to specific chemical concepts according to semantic categories (another area of interest, the semantic web and chemistry, which I will blog about later in the year).
Chemical Content Recognition
This is a service delivered by MPirics. It allows chemists to chemical entities from text, the main goal being patents.
Subscribe to:
Posts (Atom)