Since I'm now going to be coding predominately in C++, I thought it would be nice to keep the Java going and give my website a new face. Whilst I'm not going to compare different
JavaScript frameworks, I will give a few pointers as to why I chose the Google Web Toolkit (GWT).
- Firstly, I have previously developed a semantic web application using the GWT.
- Secondly, I have a stronger background in Java programming than Javascript. GWT allows you to write AJAX applications in Java and then compile the source to highly optimized JavaScript that runs across all browsers.
- Thirdly, because its Java I can use my favourtie IDE, Eclipse. Eclipse has a large number of tools to assist the developer, it also has the advantage that it comes with a GWT plugin, which can create projects seamlessly. Eclipse supports a number of version control systems (CVS, SVN, GitHub), necessary for any software project.
So if one were to develop a website or an online application, with GWT and Eclipse and support it under say the GitHub, how would one go about doing that?
Just follow these steps:
- Download Eclipse from http://www.eclipse.org/downloads/
- Add the GWT plugin. The appropriate urls are:
Eclipse 3.6 (Helios)
http://dl.google.com/eclipse/plugin/3.6
Eclipse 3.5 (Galileo)
http://dl.google.com/eclipse/plugin/3.5
Eclipse 3.4 (Ganymede)
http://dl.google.com/eclipse/plugin/3.4
Eclipse 3.3 (Europa)
http://dl.google.com/eclipse/plugin/3.3
3.
Restart your Eclipse to make sure the workspace is refreshed.
4.
To create a new GWT project, simply click on the GWT project icon in blue and white, or go to File-> New -> Project -> Google -> WebApplicationProject.
Click on the next button, then you will be prompted for a project name and package. You will also be given the option to specify the SDK you want to use in my case I have installed both the GWT -1.7.0 and GWT 2.0.4 versions. I would suggest upgrading to the newest version as from the GWT 2.0 mile stone, they no longer provide individual operating system .jar files. Previously I had a gwt-dev-linux.jar for my Ubuntu machine, however now the distributable .jar is called gwt-dev.jar.
This should produce a project structure similar to the one on my
GitHub.
Code for the main class has been copied in here:
package eoc21.client;
import eoc21.shared.FieldVerifier;
import com.google.gwt.core.client.EntryPoint;
import com.google.gwt.core.client.GWT;
import com.google.gwt.event.dom.client.ClickEvent;
import com.google.gwt.event.dom.client.ClickHandler;
import com.google.gwt.event.dom.client.KeyCodes;
import com.google.gwt.event.dom.client.KeyUpEvent;
import com.google.gwt.event.dom.client.KeyUpHandler;
import com.google.gwt.user.client.rpc.AsyncCallback;
import com.google.gwt.user.client.ui.Button;
import com.google.gwt.user.client.ui.DialogBox;
import com.google.gwt.user.client.ui.HTML;
import com.google.gwt.user.client.ui.Label;
import com.google.gwt.user.client.ui.RootPanel;
import com.google.gwt.user.client.ui.TextBox;
import com.google.gwt.user.client.ui.VerticalPanel;
/**
* Entry point classes define
onModuleLoad().
*/
public class GWTWebSite implements EntryPoint {
/**
* The message displayed to the user when the server cannot be reached or
* returns an error.
*/
private static final String SERVER_ERROR = "An error occurred while "
+ "attempting to contact the server. Please check your network "
+ "connection and try again.";
/**
* Create a remote service proxy to talk to the server-side Greeting service.
*/
private final GreetingServiceAsync greetingService = GWT
.create(GreetingService.class);
/**
* This is the entry point method.
*/
public void onModuleLoad() {
final Button sendButton = new Button("Send");
final TextBox nameField = new TextBox();
nameField.setText("GWT User");
final Label errorLabel = new Label();
// We can add style names to widgets
sendButton.addStyleName("sendButton");
// Add the nameField and sendButton to the RootPanel
// Use RootPanel.get() to get the entire body element
RootPanel.get("nameFieldContainer").add(nameField);
RootPanel.get("sendButtonContainer").add(sendButton);
RootPanel.get("errorLabelContainer").add(errorLabel);
// Focus the cursor on the name field when the app loads
nameField.setFocus(true);
nameField.selectAll();
// Create the popup dialog box
final DialogBox dialogBox = new DialogBox();
dialogBox.setText("Remote Procedure Call");
dialogBox.setAnimationEnabled(true);
final Button closeButton = new Button("Close");
// We can set the id of a widget by accessing its Element
closeButton.getElement().setId("closeButton");
final Label textToServerLabel = new Label();
final HTML serverResponseLabel = new HTML();
VerticalPanel dialogVPanel = new VerticalPanel();
dialogVPanel.addStyleName("dialogVPanel");
dialogVPanel.add(new HTML("
Sending name to the server:"));
dialogVPanel.add(textToServerLabel);
dialogVPanel.add(new HTML("
Server replies:"));
dialogVPanel.add(serverResponseLabel);
dialogVPanel.setHorizontalAlignment(VerticalPanel.ALIGN_RIGHT);
dialogVPanel.add(closeButton);
dialogBox.setWidget(dialogVPanel);
// Add a handler to close the DialogBox
closeButton.addClickHandler(new ClickHandler() {
public void onClick(ClickEvent event) {
dialogBox.hide();
sendButton.setEnabled(true);
sendButton.setFocus(true);
}
});
// Create a handler for the sendButton and nameField
class MyHandler implements ClickHandler, KeyUpHandler {
/**
* Fired when the user clicks on the sendButton.
*/
public void onClick(ClickEvent event) {
sendNameToServer();
}
/**
* Fired when the user types in the nameField.
*/
public void onKeyUp(KeyUpEvent event) {
if (event.getNativeKeyCode() == KeyCodes.KEY_ENTER) {
sendNameToServer();
}
}
/**
* Send the name from the nameField to the server and wait for a response.
*/
private void sendNameToServer() {
// First, we validate the input.
errorLabel.setText("");
String textToServer = nameField.getText();
if (!FieldVerifier.isValidName(textToServer)) {
errorLabel.setText("Please enter at least four characters");
return;
}
// Then, we send the input to the server.
sendButton.setEnabled(false);
textToServerLabel.setText(textToServer);
serverResponseLabel.setText("");
greetingService.greetServer(textToServer,
new AsyncCallback
() {
public void onFailure(Throwable caught) {
// Show the RPC error message to the user
dialogBox
.setText("Remote Procedure Call - Failure");
serverResponseLabel
.addStyleName("serverResponseLabelError");
serverResponseLabel.setHTML(SERVER_ERROR);
dialogBox.center();
closeButton.setFocus(true);
}
public void onSuccess(String result) {
dialogBox.setText("Remote Procedure Call");
serverResponseLabel
.removeStyleName("serverResponseLabelError");
serverResponseLabel.setHTML(result);
dialogBox.center();
closeButton.setFocus(true);
}
});
}
}
// Add a handler to send the name to the server
MyHandler handler = new MyHandler();
sendButton.addClickHandler(handler);
nameField.addKeyUpHandler(handler);
}
}
Essentially we create a class for the entry point for the website (i.e. home), we implement the onModuleLoad() method. The tutorial defines a number of widgets: a button called "send", a TextBox with a default value "GWT", the text box and button are added to the root panel. A DialogBox is then created that will display the results. A close button is added to the DialogBox with a click handler to close the DialogBox if a user clicks on the close button.
A nested class called MyHandler is constructed that implements both the ClickHandler and KeyUpHandler interfaces, which has to implement the onClick() and onKeyUpUp() methods. If a user clicks the send button or types in information in the textbox, it is sent to the server, via the sendNameToServer() method, which implements an asynchronous remote proceedual call to the server.
5. Get Egit
You're probably now wondering how I integrated Eclipse with GitHub? Well, there is also a plugin to allow you to push your code to GitHub. The plugin is called Egit, and can be downloaded from: http://download.eclipse.org/egit/updates. A full and more comprehensive tutorial on Egit can be found at http://www.vogella.de/articles/EGit/article.html.
6. Mavenize and manage those dependencies
If you're new to Java, or you haven't heard of Maven, you should read this (http://maven.apache.org/). Apache Maven is a software project management and comprehension tool. Based on the concept of a project object model (POM), Maven can manage a project's build, reporting and documentation from a central piece of information. I will talk about maven, and mavenizing a GWT project in another post.
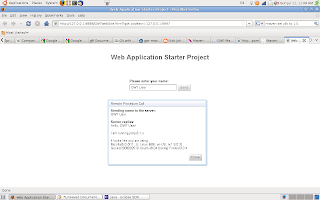
GWT Simple Demo
The result of our simple GWT application / website demo is shown in the image below. On clicking the send button an RPC (remote proceedual call) is made, the server responds with "Sending name to the server"+ your input text, then it replies with "hello" and the name of the user and details of the operating system and browser used (in this case Ubuntu 8.0.4 and FireFox).












